|
Polygonal random fields are a natural model for robotic mapping problems
where we must classify each point in the environment as free space or as
occupied by some object. In these problems we typically get observations
from laser or sonar rangefinders. An example is shown below.

(a) localized laser data |

(b) occupancy grid |
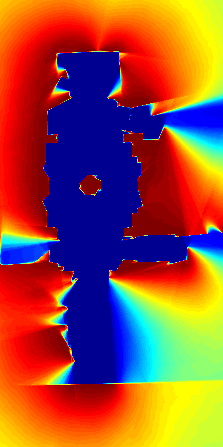
(c) polygonal random field posterior |
 |
| Figure: an example of
robotic mapping with polygonal random fields. (a) A data set of 5,500
localized laser observations. (b) An occupancy grid computed from the
data. (c) A visualization of the posterior distribution of a polygonal
random field. In (b) and (c) hotter colors indicate more likely occupied,
and cooler colors indicate more likely free space. (Using colors rather
than shades of gray makes it easier to see subtle differences in
probability.) |
The traditional technique for mapping with rangefinder data is occupancy
grids, which treat the environment as a grid of independent cells that are
either occupied or free. Polygonal random fields yield improved maps
because they have spatial correlation—nearby points are likely to
have the same colors—and they are biased towards colorings with
planar boundaries, which are prevalent in many mapping problems. This
bias leads to useful extrapolation, as at the bottom of the map, where the
set of collinear laser impacts has provided evidence for a wall; the
polygonal random field extrapolates that this wall extends for some
unknown distance.
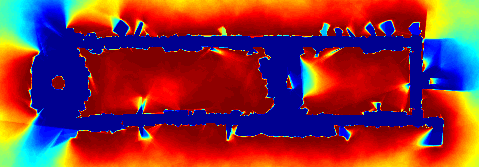
Another example is shown below, this time with sonar data. Sonar data
are far more difficult to interpret than laser data; while a laser impact
indicates a point of contact, a sonar impact yields an arc which
contains a point of impact. Because sonar data is more ambiguous,
occupancy grids perform poorly with sonar data. Many of the walls are
missing due to specular reflections, and the walls that remain are
indistinct. In this context, polygonal random fields yield far better
maps: the boundaries are crisp, and the free space is clearly marked.
Surprisingly, the technique also correctly identifies a number of rooms
using a combination of subtle cues: a small number of longer range
readings that enter the room, and corner impacts at the door frame.
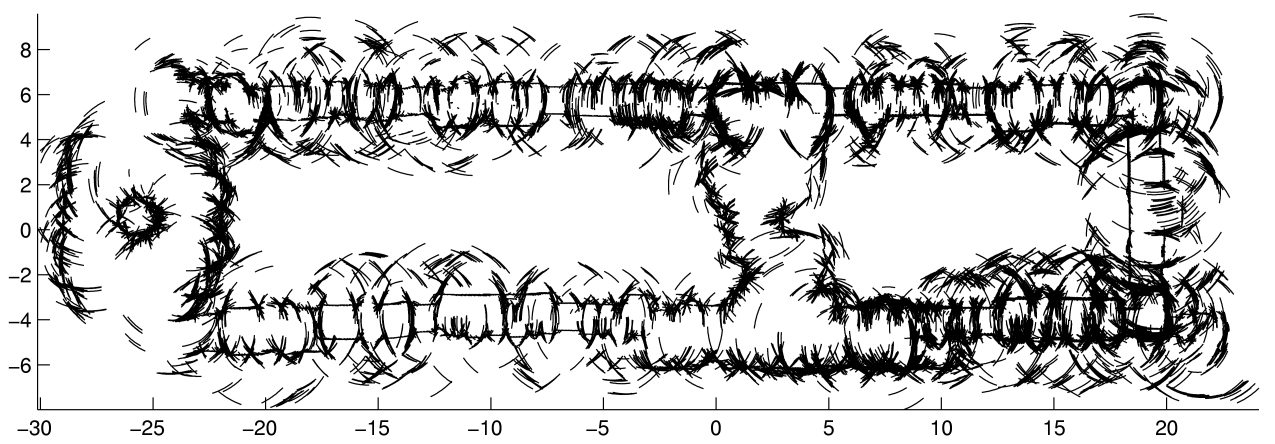
(a) localized sonar data |
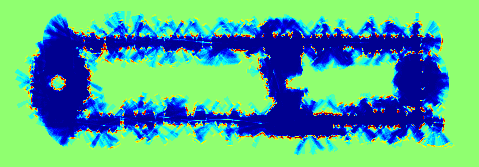
(b) occupancy grid |
(c) polygonal random field posterior |
| Figure: an example of
sonar mapping with polygonal random fields. (a) A data set of 29,120
localized sonar observations. (b) An occupancy grid computed from the
data. (c) A visualization of the posterior distribution of a polygonal
random field. In (b) and (c) hotter colors indicate more likely occupied,
and cooler colors indicate more likely free space. (Using colors rather
than shades of gray makes it easier to see subtle differences in
probability.) |
A movie (6MB) that shows some samples drawn
from the posterior is also available. Note how in the regions with dense
observations, the posterior is very peaked; in regions without many
measurements the sampler explores alternative structures for the environment.
|
|
The paper is available for download in PDF format.
@InProceedings{PaskinThrun2005,
author = {Mark A. Paskin and Sebastian Thrun},
title = {Robotic Mapping with Polygonal Random Fields},
booktitle = {Proceedings of the 21st Conference on
Uncertainty in Artificial Intelligence},
pages = {??--??},
year = {2005},
editor = {Faheim Bacchus and Tommi Jaakkola},
month = {July},
publisher = {AUAU Press, Arlington, Virginia}
}
|
|
licensing
This implementation is © Mark A. Paskin 2005. I am distributing
my implementation as open-source software under the GNU Public License.
This distribution should build on *nix systems without a problem, and
porting it to Windows should not be difficult. I developed it on Linux
and Mac OS X.
disclaimer
I am making this code available so that the experiments in the paper
are reproducible, and to facilitate research on applications of
polygonal random fields. (This implementation took me months to
develop, and I'd like other researchers to be able to avoid this startup
cost.) However, the code is not supported, so if you plan to use
it please prepare to get your hands dirty. If you have trouble with my
software (but not with CGAL or Qt) I am happy to answer questions via
e-mail.
installation
Below are the steps to installing this distribution:
Install the GNU
Scientific Library (GSL). Install Qt with
support for multi-threading. Qt is a library for developing C++
graphical user interfaces, and it is used by the visualization
software. An open-source
distribution of Qt is available. My distribution requires multiple
threads to simultaneously perform sampler computations and visualize
the state of the sampler. Install CGAL with support for
Qt. CGAL is a wonderful open-source library for geometric
computation, and it is used extensively in my implementation for
geometric primitives like points and line segments, and algorithms
like intersection testing. Download my distribution, arak.zip(1 MB), and unzip
it.
Make sure that the environment variable CGAL_MAKEFILE
is defined; this should have been taken care of when you installed
CGAL. cd into the directory arak and type
make. To make the documentation:
- Install Doxygen, an open-source source code documentation program.
- Type make doc.
getting started
The main program is bin/sampler, which uses MCMC to sample
from several types of Arak processes. Below I'll step through an
example of sampling from an Arak process with Gaussian observations.
- Take a look at the file data/gaussian.props. This file
contains a number of properties that govern how the sampler operates.
There are parameters which govern:
- the boundary of the observation window;
- the scale parameter of the Arak process;
- the type of observations (Gaussian in this case);
- the parameters of the observation model;
- a path to the file containing the observed data;
- parameters of the Clifford & Nicholls (1994) proposal distribution;
- parameters of the Markov chain, including burn-in, stride, and annealing;
- parameters of the color estimates to compute; and
- paths to where the results are written, including the final coloring, the color estimates, the most likely coloring found, and a subset of the samples from the Markov chain.
Run the sample using these properties by typing
bin/sampler -v -t -p data/gaussian.props
The -v flag requests visualization of the sampler's state,
and the -t flag requests a navigation toolbar in the GUI.
The -p option takes a path to a file containing the
parameters of the simulation. These parameters can be specified
explicitly (or overridden) on the command line using the -D
option, as in
bin/sampler -v -t -p data/gaussian.props -D arak.scale=2.0
which overrides the Arak process scale parameter to be 2.0.
The first thing you will see is the sampler write out the parameter
values of the simulation. Then a window should pop up, showing the
current state of the sampler. It will look something like this:
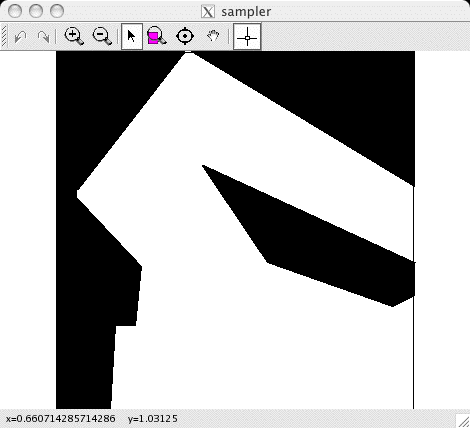
The toolbar at the top allows you to zoom and pan the display. It
also stores a history of these actions, which can be reversed using
the left and right arrows (on the left side of the toolbar).
Pressing the right mouse button will cycle through this view and
three other views:
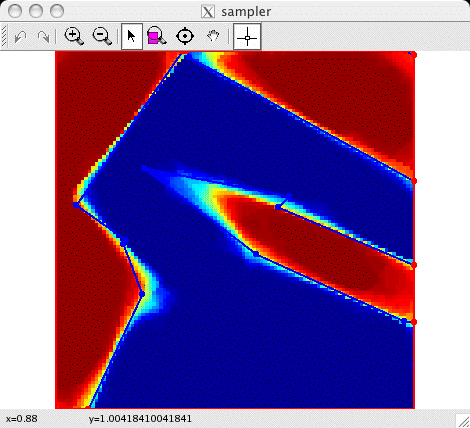 |
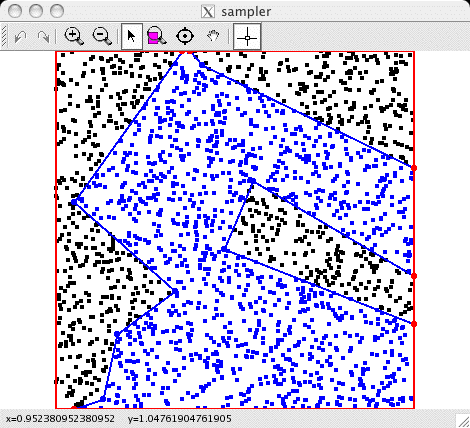 |
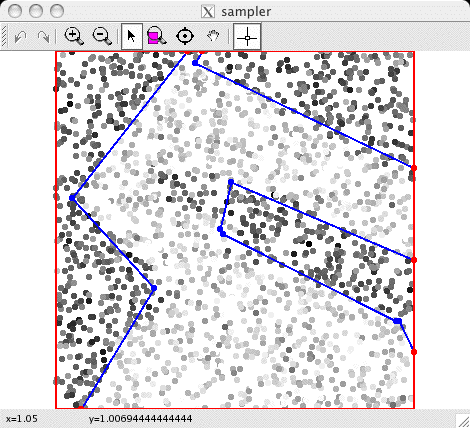 |
| The color estimates for a grid of point
locations. Each color estimate is the probability a point is
colored black; this probability is visualized on a color scale
(identical to the Matlab "JET" colormap) where dark blue
corresponds to zero and dark red corresponds to one. |
The current colors of the sampler's query points.
These are the points whose colors are maintained explicitly by the
sampler for efficiency; in this example, the points correspond to
the locations of observations so likelihoods can be evaluated
quickly. |
A visualization of the observed data, with the
current discontinuity graph overlaid. |
Quit the sampler by closing the visualization window. The result
files are written out and some statistics about the Markov chain (like
acceptance ratios for different move types) are written to the shell.
When the visualization is not requested (by omitting the -v
flag), the sampler can be stopped by typing Control-C (or signalling
INT). Run the sampler using other types of observations by changing the
property file you use:
| data/gaussian.props | The
Gaussian data used in the original Clifford & Nicholls (1994)
paper. |
| data/gaussian-circle.props |
Some Gaussian data generated by a black circle in an otherwise
white plane. It is interesting to see how averaging over polygonal
colorings can yield a smooth circular image. |
| data/bernoulli.props | Some
boolean data corrupted by bit-flipping noise. This shows how Arak
processes could be used to perform anisotropic smoothing of bitmap
images. |
| data/prf-demo.props | Another
anisotropic smoothing example; this was used to generate the movie
at the top of this page. |
| data/range.props | This is some
simulated range data generated using Player/Stage. For
range data the data set is visualized by drawing each range
measurement as a line segment from the sensor location to the impact
point; the color of this line segment indicates its likelihood under
given the current coloring. |
| data/gates-laser.10.lobby.props | This
is 1/10th of a laser data gathered in the lobby of the Gates
Building at Stanford. It consists of around 5,500 localized laser
rangefinder measurements. This is the laser rangefinder data set
reported in the UAI paper. |
| data/gates-laser.100.props | This is
1/100th of the complete laser dataset from the A wing of the Gates
Building at Stanford. It consists of around 3,300 localized laser
rangefinder measurements. |
| data/gates-sonar.props | This is
sonar dataset from the A wing of the Gates Building at Stanford. It
consists of around 29,000 localized sonar rangefinder measurements.
This is the sonar rangefinder data set reported in the UAI
paper. |
Note that for the more complex data sets, the computation required
for visualization can be quite large. To get faster sampling, turn
visualization off.
digging deeper...
The sampler program is the main program in this
distribution, but there are others which you may find useful.
bin/sampler writes out colorings using a simple
(self-documenting) text format. For example, if the property file
requests it, the sampler writes out the last state of the Markov chain
and/or the most likely state visited. You can use
bin/coloringtofig to convert this format to FIG format, so
that you can use xfig to view the coloring.
bin/coloringtofig reads the coloring from standard input and
writes the fig file to standard output. You can also use the
shell script bin/coloringtopdf (which uses
bin/coloringtofig internally) to create a PDF rendering of
the coloring. Finally, the script vc is a quick way to take
a look at a coloring: it invokes coloringtopdf to create a
temporary PDF rendering, and then invokes ggv to show
it. bin/sampler also writes out colorings in a more
compressed binary format to record a subset of samples from a Markov
chain simulation. To visualize these samples, you can use the program
bin/samplestofigs to convert the file of binary samples to
FIG files using
bin/samplestofigs --prefix sample- < samples.bin
This will create sample-1.fig, sample-2.fig, and so
on. I used fig2dev to convert these FIG files to PNG files,
and then used Quicktime to generate movies depicting the evolution of
the Markov chain. (You can also use the --stride option to
skip samples.)
Besides colorings, bin/sampler also writes out point
color estimates. (ogrid, described below, also does this.)
To visualized these point color estimates, there are programs which
create PGM images (where grayscale is used to visualize the
probability each point is black) and PPM images (which use the JET
colormap to map black probabilities to colors). The main programs to
use for this purpose are gesttopgm and gesttoppm
(gest = grid of color estimates). For example, if you run
bin/sampler and it writes out color estimates to
estimates.out.gz, you can run
bin/gesttoppm estimates.out.gz
to create a PPM visualization called estimates.ppm.
gesttopgm and gesttoppm are shell scripts that wrap
two programs called gridtopgm and gridtoppm which do
all the work. (There are also two programs, surftopgm and
esttopgm which use interpolation to generate PGM renderings
from point estimates that are at arbitrary locations, rather than on a
grid. They do not operate on estimates generated by
bin/sampler; I've included them because I used them at an
earlier stage of development, and they might be useful to others.)
bin/sonar_viz is a program that can be used to
visualize the sonar observation model described in the UAI paper. I
used it to design the sonar model and to choose reasonable parameters
for it. The program requires a properties file specified with a
-p option, and a fixed coloring, read from standard input.
For example, if coloring.out is a coloring generated from
bin/sampler using the properties file
data/gates-sonar.props, then you can run
bin/sonar_viz -p data/gates-sonar.props < coloring.out
to visualize the sonar observation model described by the parameters
in data/gates-sonar.props. When the window comes up, it
should show the coloring supplied as input. (If not, resize the
window and it will show up.) Click on a point in free space, and then
click on another to define a ray; this ray gives the location and
orientation of a hypothetical sonar sensor. At the bottom of the
screen you will see the likelihood function associated with range
measurements from that sensor, given the coloring is the true map. An
example is shown below (I zoomed in on a part of the map):
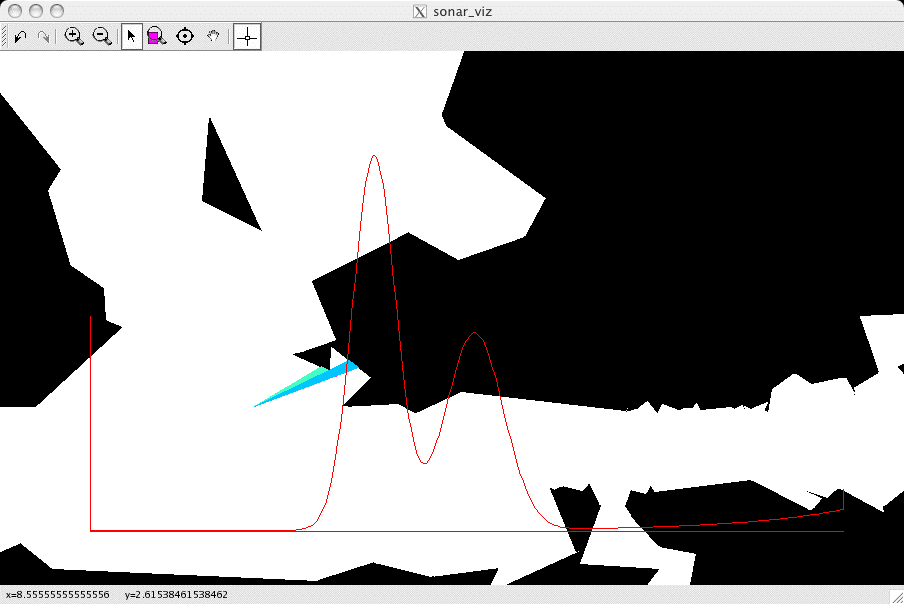
The visible faces and corners are drawn into the map, and color is
used to indicate the probability that a return would be generated by
each such feature. In the example above, there are two faces visible
to the sensor, and the closer one is more likely to generate the
return (the colormap used here is jet, as above). At the bottom you
can see the likelihood function (which has two strong modes, one for
each feature visible to the sonar sensor. ogrid is a program which computes occupancy grid
estimates associated with laser and sonar range data. Like
sampler, ogrid uses property files to determine the
input data and model parameters; see
data/gates-sonar.occupancy.props for an example which can
generate occupancy grids for the same data set as used in
data/gates-sonar.props. There are parameters which govern:
- the boundary of the occupancy grid and its resolution;
- the parameters of the inverse sensor model; and
- a path to the file containing the observed data.
The program writes out the result to standard output in the same
"grid"; format that sampler uses to write the grid
of point color estimates (although it is not gzipped). You
can therefore run
bin/ogrid -p data/gates-sonar.occupancy.props | gzip >! ogrid-estimates.out.gz
to get the same kind of estimates written out by sampler.
Then you can run gridtoppm as above to generate a
visualization.
development
I have tried to keep the code clean and well-documented. The
auto-generated documentation is available here, thanks to Doxygen.
acknowledgments
Thanks go to
- Brian Gerkey, who
helped me collect the laser and sonar data from Gates Hall
- Andrew Howard,
whose pmap library
was used to localize these range measurements.
Also, thanks go to the developers of the following pieces of software,
which are included (in whole or in part) in this distribution:
And finally, thanks to the developers of CGAL for writing a wonderful
library!
|











