Home
Search
Browse results Datasets
Figures
Clinical annotations
Overview of results GeneXPress
GeneSets
Links
People
Supplemental Information
|
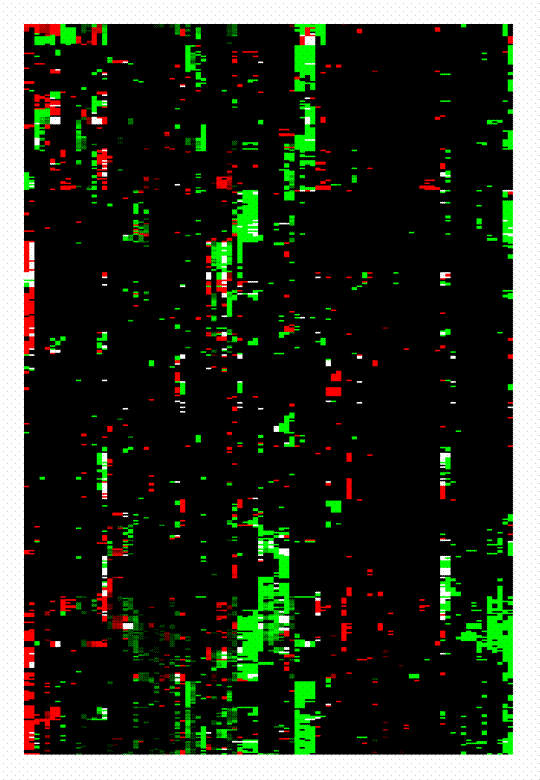
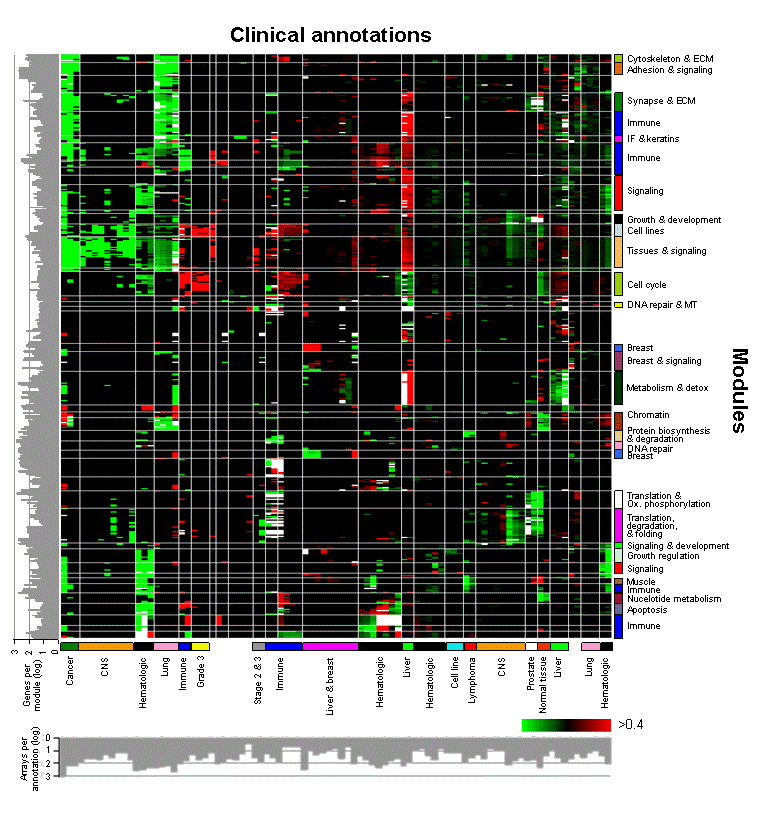
A 'module map' showing conditional activity of expression modules in cancer |
|||||||||||||||
|
|||||||||||||||
|
DNA microarrays are widely used to study the gene expression changes in tumors. These studies, however, are typically system-specific, and do not address the commonalities and variations between different types of tumor. We present an integrated analysis of a compendium of 1975 published microarrays spanning 22 tumor types. To understand this wealth of data, we describe expression profiles in different tumors in terms of the behavior of modules, sets of genes that act in concert to perform a specific function. Using a simple unified analysis, we extract modules and characterize tumor expression profiles as a combination of activated and deactivated modules. Activation of some modules is highly specific to particular types of tumor. For example, we found a growth-inhibitory module, which is specifically repressed in acute lymphoblastic leukemias, and may underlie the deregulated proliferation in these cancers. Other modules are shared across a diverse set of clinical conditions, suggesting common tumor progression mechanisms. For example, the bone osteoblastic module spans a variety of tumor types and includes both secreted growth factors and their receptors. This finding suggests a single mechanism for both primary tumor proliferation and metastasis to bone. Our analysis provides a comprehensive roadmap for cancer, offering multiple research directions for diagnostic, prognostic and therapeutic studies.
|
|||||||||||||||
 |
|||||||||||||||
|
Questions and comments regarding this web site should be addressed to
Eran Segal
This material is based upon work supported by the National Science Foundation under Grant No. 0345474, and by NIGMS. Any opinions, findings, and conclusions or recommendations in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. |
|||||||||||||||