| | ||||
 |
|
|||
 >4x >4x |
Module 152 -- Oxidative phosphorylation and ATP synthesis |
| General Properties | |
|---|---|
| Below we list the number of gene sets from which the module was composed was originally composed, and the number of genes and experiments that it contains. Clicking on the number of genes will display a detailed list of all the genes in the module. Clicking on the number of experiments will display a detailed list of all the experiments in the module. | |
| Num gene sets: | 23 |
| Num genes: | 124 (see 1706 additional genes for this module) |
| Num experiments: | 291 (135 induced, 156 repressed) |
| Parent module: | None |
| Children modules: | 62 103 77 116 |
| Gene sets | |
|---|---|
| Below we list the gene sets from which the module was originally composed. Clicking on a gene set will display all the genes associated with that gene set. Also listed are all the other modules in which the gene set participates. | |
| nucleoside monophosphate phosphorylation | |
| ATP_Synthesis | |
| proton transport | |
| mitochondrial inner membrane | |
| cytochrome c oxidase activity | |
| cbb3_type cytochrome c oxidase | |
| NADH dehydrogenase activity | |
| NADH dehydrogenase (ubiquinone) activity | |
| Ubiquinone_biosynthesis | |
| protein_disulfide reduction | |
| mitochondrial electron transport, NADH to ubiquinone | |
| ATP synthesis coupled electron transport (sensu Eukarya) | |
| oxidative phosphorylation | |
| mitochondrion | |
| ion transporter activity | |
| mitochondrial membrane | |
| inner membrane | |
| hydrogen ion transporter activity | |
| Oxidative_phosphorylation | |
| monovalent inorganic cation transporter activity | |
| sodium ion transporter activity | |
| metal ion transporter activity | |
| Electron Transport Chain | |
| Enriched clinical annotations | |||||||
|---|---|---|---|---|---|---|---|
| For each of the clinical annotations, we tested whether it was enriched in the set of arrays in which the module is significantly induced (or repressed). Below we list all such annotations that were enriched in this module with an FDR corrected p-value of 0.05 ('P-value' column), along with the number ('Hits' column) of induced (or repressed) module's arrays in which they appear. We also show the fraction of the module's induced (or repressed) arrays with the given annotation ('Hits (%)' column). For completeness, we also list the total number of relevant induced arrays in the module ('Module hits in category' column), total number of arrays in the compendium in which the corresponding annotation is present ('Arrays in annotation' column), and the total number of relevant arrays in the compendium ('Arrays' column). Note that the 'relevant' arrays may be different for each annotation, since it includes only arrays in which the annotation was relevant (i.e., present or not present) and excludes arrays where the value of the annotation was not known. Rows that correspond to annotations that were enriched in the induced arrays of the module have a red background, while rows corresponding to annotations enriched in the repressed arrays have a green background. For each annotation, we also list the other modules in which it was enriched. | |||||||
| Clinical annotation | Category | P-value | Hits | Hits(%) | Module hits in category | Arrays in annotation | Arrays |
| Prostate (Prostate cancer*) | Prostate cancer* | 3.5e-21 | 41 | 26.4 | 155 | 101 | 1945 |
| Hematologic cancer and cell line (Stimulated immune*) | Stimulated immune* | 4.8e-11 | 20 | 14.8 | 135 | 53 | 1945 |
| Hematologic samples and cell lines (Stimulated immune*) | Stimulated immune* | 4.8e-11 | 20 | 14.8 | 135 | 53 | 1945 |
| Macrophages (Stimulated immune*) | Stimulated immune* | 4.8e-11 | 20 | 14.8 | 135 | 53 | 1945 |
| Monocytes (Stimulated immune*) | Stimulated immune* | 4.8e-11 | 20 | 14.8 | 135 | 53 | 1945 |
| Lung carcinoid (Lung cancer) | Lung cancer | 2.9e-08 | 9 | 60 | 15 | 20 | 276 |
| CNS cancer or cell line (Neuro tumors*) | Neuro tumors* | 1.0e-07 | 22 | 14.1 | 155 | 81 | 1945 |
| CNS tumor (Neuro tumors*) | Neuro tumors* | 1.0e-07 | 22 | 14.1 | 155 | 81 | 1945 |
| CNS tissue, cancer or cell line (Neuro tumors*) | Neuro tumors* | 2.5e-07 | 22 | 14.1 | 155 | 85 | 1945 |
| Cancer and cell line (Neuro tumors*) | Neuro tumors* | 3.2e-07 | 22 | 14.1 | 155 | 86 | 1945 |
| Cancer (Neuro tumors*) | Neuro tumors* | 3.2e-07 | 22 | 14.1 | 155 | 86 | 1945 |
| Hematologic cancer and cell line (Stimulated immune*) | Stimulated immune* | 7.4e-06 | 15 | 9.6 | 155 | 53 | 1945 |
| Hematologic samples and cell lines (Stimulated immune*) | Stimulated immune* | 7.4e-06 | 15 | 9.6 | 155 | 53 | 1945 |
| Macrophages (Stimulated immune*) | Stimulated immune* | 7.4e-06 | 15 | 9.6 | 155 | 53 | 1945 |
| Monocytes (Stimulated immune*) | Stimulated immune* | 7.4e-06 | 15 | 9.6 | 155 | 53 | 1945 |
| Hematologic cancer and cell line (Stimulated PBMC) | Stimulated PBMC | 1.2e-05 | 6 | 100 | 6 | 30 | 182 |
| Macrophages (Stimulated PBMC) | Stimulated PBMC | 1.2e-05 | 6 | 100 | 6 | 30 | 182 |
| Metastasis (Liver cancer) | Liver cancer | 6.2e-05 | 6 | 30 | 20 | 10 | 207 |
| Follicular lymphoma (B lymphoma) | B lymphoma | 0.0001 | 8 | 47.0 | 17 | 28 | 245 |
| Follicular lymphoma (Various tumors) | Various tumors | 0.0001 | 5 | 38.4 | 13 | 9 | 154 |
| Enriched GO annotations | ||||||
|---|---|---|---|---|---|---|
| For each GO annotation, we tested whether it was enriched in the set of genes of the module. Below we list all such annotations that were enriched in this module with an FDR corrected p-value of 0.05 ('P-value' column), along with the number ('Hits' column) of the module's genes in which they appear. We also show the fraction of the module's genes with the given GO annotation that are included in this module ('Hits (%)' column). For completeness, we also list the total number of genes in the module ('Module genes' column), total number of genes in the compendium in which the corresponding annotation is present ('Genes in annotation' column), and the total number of genes in the compendium ('Genes' column). | ||||||
| GO annotation | P-value | Hits | Hits(%) | Module genes | Genes in annotation | Genes |
| mitochondrion | 1.3e-134 | 103 | 88.0 | 117 | 207 | 4566 |
| monovalent inorganic cation transporter activity | 4.4e-88 | 61 | 52.1 | 117 | 80 | 4566 |
| ion transporter activity | 3.1e-72 | 65 | 55.5 | 117 | 144 | 4566 |
| sodium ion transporter activity | 1.1e-57 | 40 | 34.1 | 117 | 50 | 4566 |
| metal ion transporter activity | 2.6e-56 | 42 | 35.8 | 117 | 60 | 4566 |
| hydrogen ion transporter activity | 4.4e-53 | 38 | 32.4 | 117 | 50 | 4566 |
| mitochondrial membrane | 8.2e-41 | 33 | 28.2 | 117 | 54 | 4566 |
| electron transporter activity | 4.2e-38 | 55 | 47.0 | 117 | 269 | 4566 |
| inner membrane | 2.4e-37 | 25 | 21.3 | 117 | 29 | 4566 |
| oxidoreductase activity | 8.7e-36 | 52 | 44.4 | 117 | 254 | 4566 |
| NADH dehydrogenase (ubiquinone) activity | 6.3e-35 | 23 | 19.6 | 117 | 26 | 4566 |
| NADH dehydrogenase activity | 2.7e-33 | 22 | 18.8 | 117 | 25 | 4566 |
| proton transport | 1.4e-27 | 22 | 18.8 | 117 | 35 | 4566 |
| mitochondrial inner membrane | 3.6e-27 | 23 | 19.6 | 117 | 41 | 4566 |
| electron transport | 3.0e-24 | 32 | 27.3 | 117 | 128 | 4566 |
| energy pathways | 4.0e-23 | 37 | 31.6 | 117 | 200 | 4566 |
| cation transporter activity | 1.3e-22 | 21 | 17.9 | 117 | 45 | 4566 |
| photosynthesis | 6.1e-22 | 33 | 28.2 | 117 | 162 | 4566 |
| oxidative phosphorylation | 1.5e-20 | 14 | 11.9 | 117 | 17 | 4566 |
| nucleoside monophosphate phosphorylation | 6.7e-20 | 14 | 11.9 | 117 | 18 | 4566 |
| monovalent inorganic cation transport | 4.1e-19 | 23 | 19.6 | 117 | 79 | 4566 |
| cytochrome c oxidase activity | 5.7e-19 | 12 | 10.2 | 117 | 13 | 4566 |
| phosphorus metabolism | 1.5e-18 | 28 | 23.9 | 117 | 138 | 4566 |
| ATP synthesis coupled electron transport (sensu Eukarya) | 1.9e-17 | 12 | 10.2 | 117 | 15 | 4566 |
| cbb3-type cytochrome c oxidase | 2.2e-17 | 11 | 9.4 | 117 | 12 | 4566 |
| phosphate metabolism | 2.7e-17 | 28 | 23.9 | 117 | 153 | 4566 |
| cation transport | 7.0e-17 | 25 | 21.3 | 117 | 120 | 4566 |
| protein-disulfide reduction | 7.5e-17 | 12 | 10.2 | 117 | 16 | 4566 |
| mitochondrial electron transport\, NADH to ubiquinone | 6.6e-16 | 11 | 9.4 | 117 | 14 | 4566 |
| phosphorylation | 1.2e-15 | 28 | 23.9 | 117 | 176 | 4566 |
| proton-transporting ATP synthase complex (sensu Eukarya) | 3.4e-14 | 9 | 7.6 | 117 | 10 | 4566 |
| photosynthesis\, light reaction | 4.2e-14 | 22 | 18.8 | 117 | 115 | 4566 |
| ion transport | 4.9e-13 | 26 | 22.2 | 117 | 188 | 4566 |
| mitochondrial electron transport chain | 1.2e-12 | 8 | 6.8 | 117 | 9 | 4566 |
| vacuolar hydrogen-transporting ATPase | 6.3e-12 | 8 | 6.8 | 117 | 10 | 4566 |
| nucleotide metabolism | 3.0e-11 | 15 | 12.8 | 117 | 64 | 4566 |
| ubiquinol-cytochrome c reductase activity | 1.0e-08 | 5 | 4.2 | 117 | 5 | 4566 |
| membrane fraction | 2.5e-08 | 26 | 22.2 | 117 | 304 | 4566 |
| outer membrane | 6.3e-07 | 6 | 5.1 | 117 | 14 | 4566 |
| ammonia oxidation | 2.0e-06 | 4 | 3.4 | 117 | 5 | 4566 |
| hydrogen-exporting ATPase activity\, phosphorylative mechanism | 5.9e-06 | 4 | 3.4 | 117 | 6 | 4566 |
| tricarboxylic acid cycle | 2.4e-05 | 5 | 4.2 | 117 | 15 | 4566 |
| ATP-binding and phosphorylation-dependent chloride channel activity | 6.4e-05 | 3 | 2.5 | 117 | 4 | 4566 |
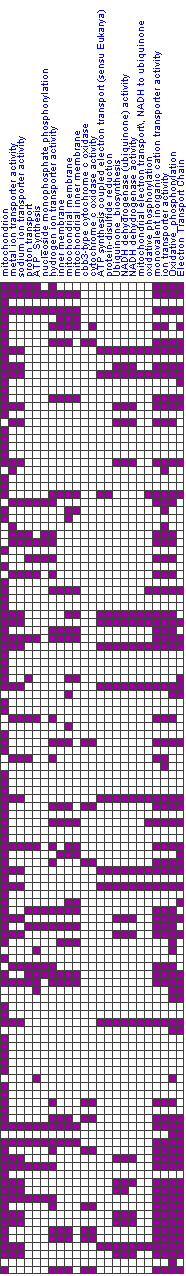
| Visual display | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Below is a visual display of the module. Shown are those arrays in which the module's genes significantly change, and the direction of change (induction/repression) in each array is indicated (middle, labeled 'Module changes'; red/green row). The arrays that correspond to the significant clinical attributes are also shown (top; brown rows). The gene sets that compose the module are also shown (left) along with an indication of the membership of the module genes in these gene sets. | |||||||||
| You can also view the images within GeneXPress by loading the module gxp file file. | |||||||||
|
|||||||||